Instructions for Using the Database
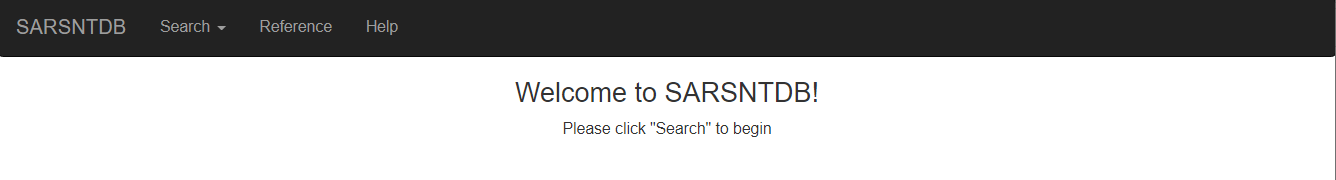
Once one accesses the database, the home screen should look as portrayed below with 4 different tabs including “Home”, “Search”, “Reference”, and “Help”.
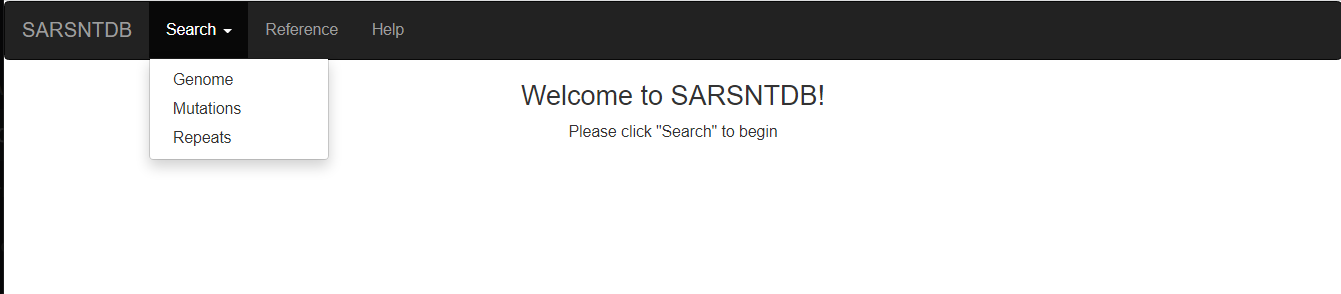
By moving the cursor over the “Search” tab, a drop-down menu will appear (as shown below). Right-click on the Genome option to be directed to where specific regions of SARS-CoV-2 can be searched (note: use the nucleotide coordinates) or one can look up a specific Gene/Protein.
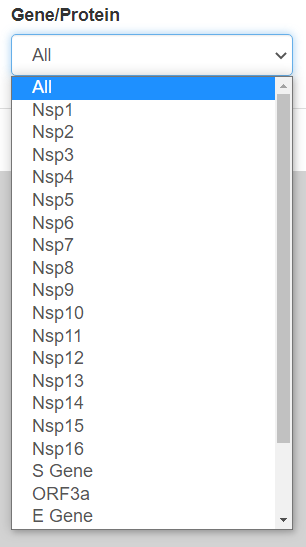
By pressing the tab that initially says “All” under Gene/Protein, a drop-down menu will appear with all the shared genes/proteins in both SARS-CoV-2 and SARS-CoV as shown below
Once you pick a specific viral component, only minimal data will appear about the gene, its encoded protein and known structural domains, accession code, specific coordinates, and brief descriptions. For more information, right-click "View Detail"
As a result, a page will appear with an image of the predicted structure of the full protein, its accepted function, information about known structural domains, and RNA and AA sequences. The page for the S gene should look as shown below
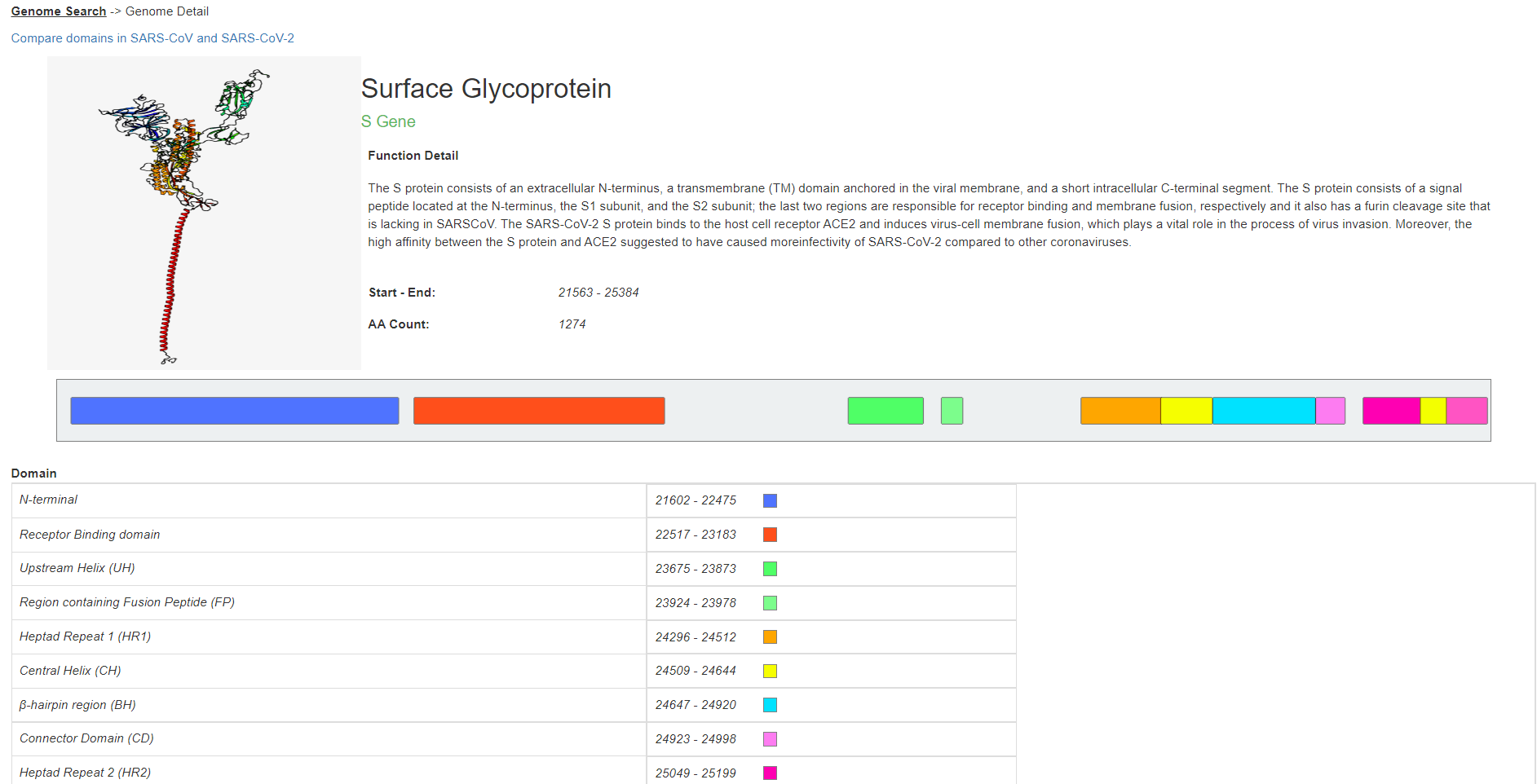
In addition, to see more information about the specific comparison between the domains in SARS-CoV and SARS-CoV-2, one can press the “Compare domains in SARS-CoV and SARS-CoV-2” button above the provided image. This will lead to a page with a description of each known domain and how it compares between the two viral strains. Each domain is color-coded and represented in its amino acid sequence according to its assigned color below the table.
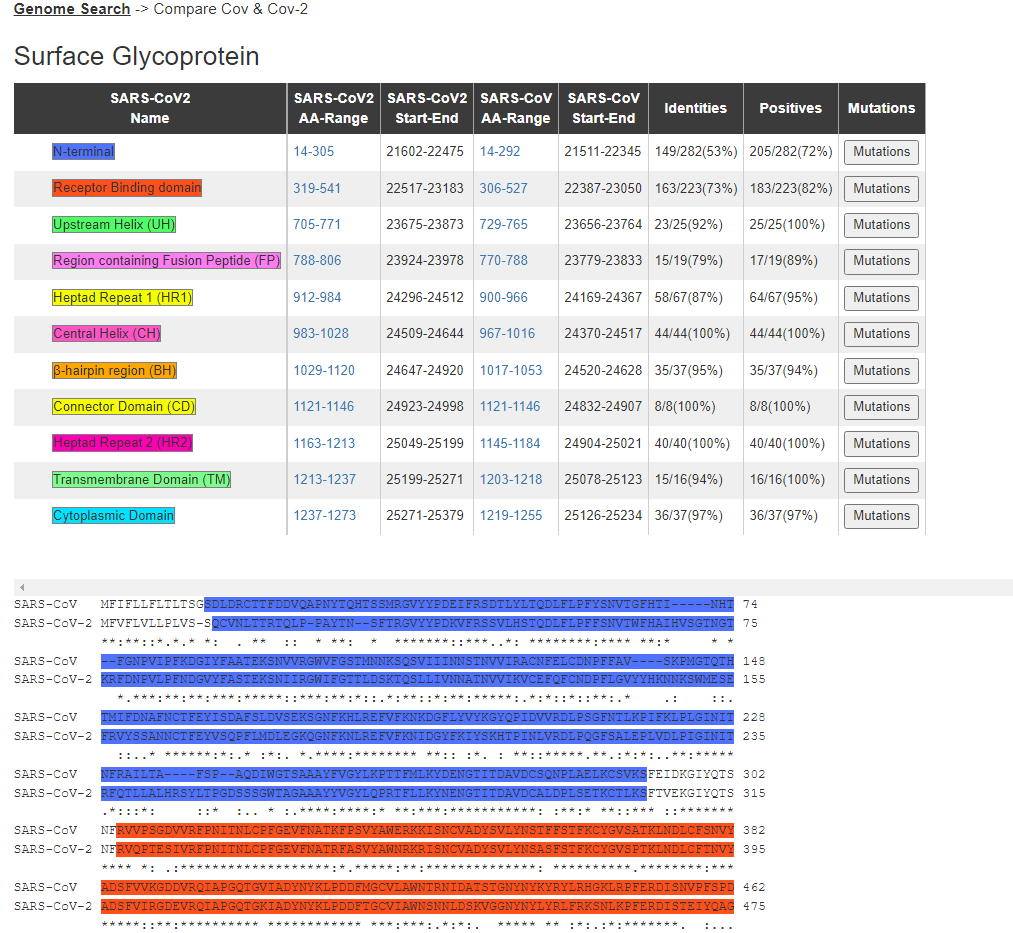
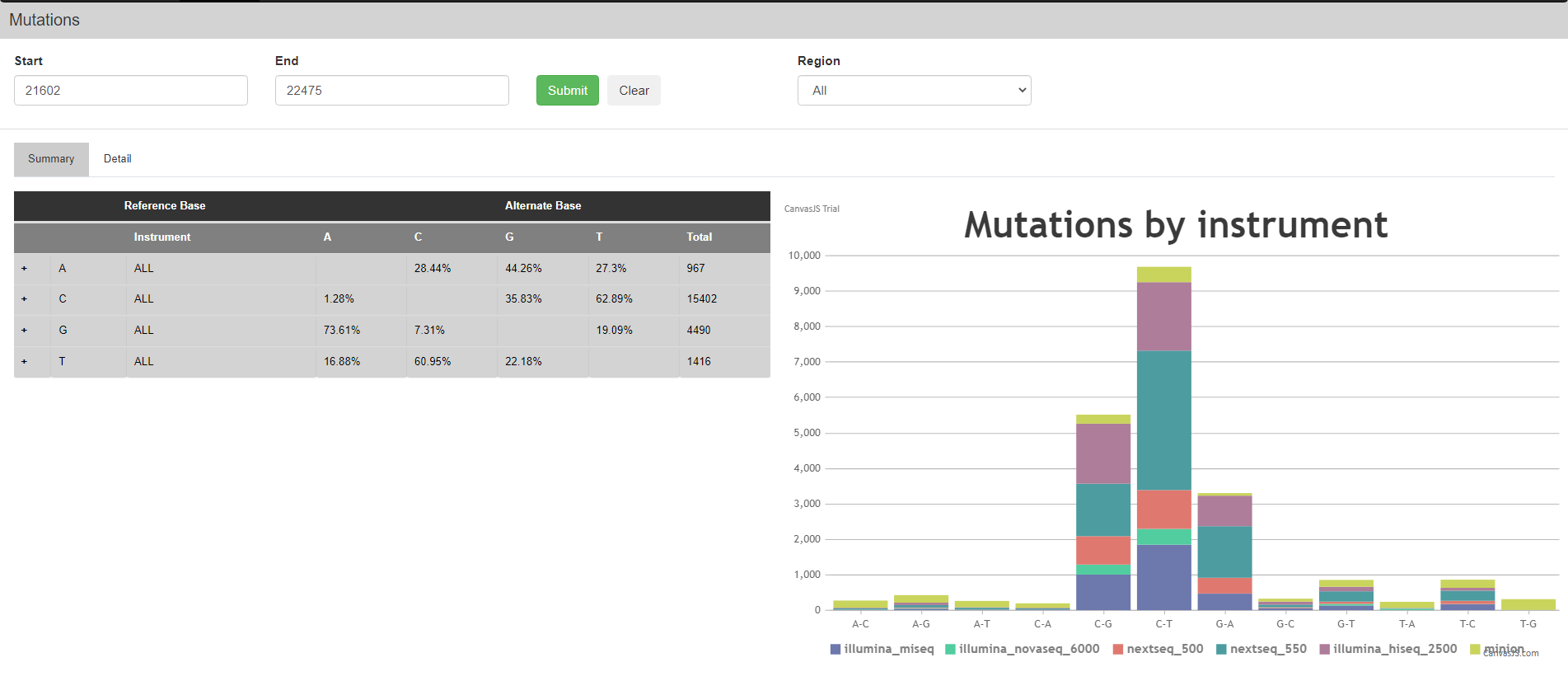
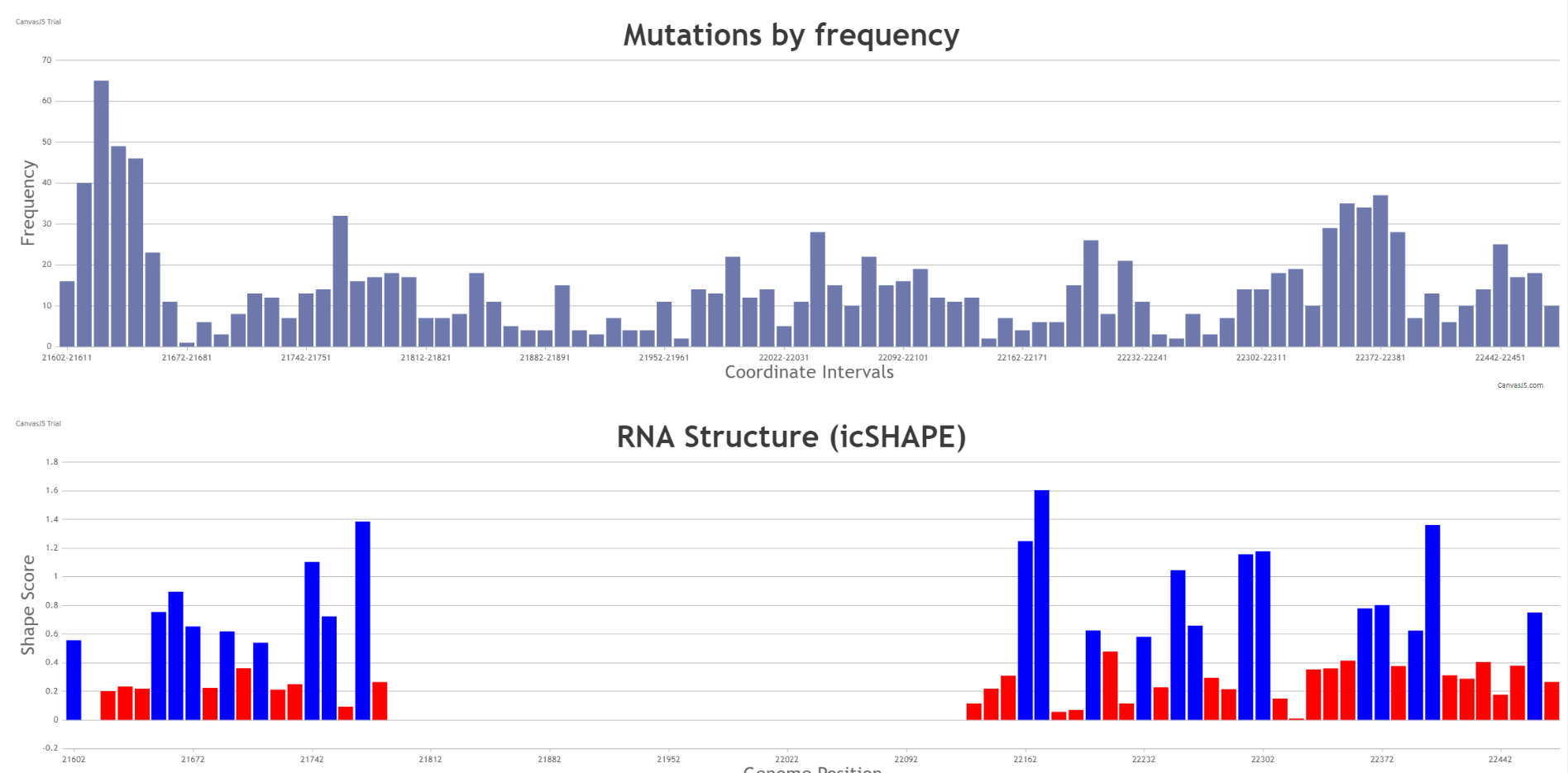
Within the table, a “Mutations” button is present in the right most column for each structural region. Once pressed, the user is redirected to the specific Mutations page that corresponds to the viral nucleotide coordinate of the corresponding structural domain of interest. The Mutations page, which is one of the options of the “Search” tab seen in the first image above, can provide a summary of all the likelihood of certain base mutations, its known frequency and RNA Structure based on icSHAPE software as shown below for the N-terminal domain of the S gene.
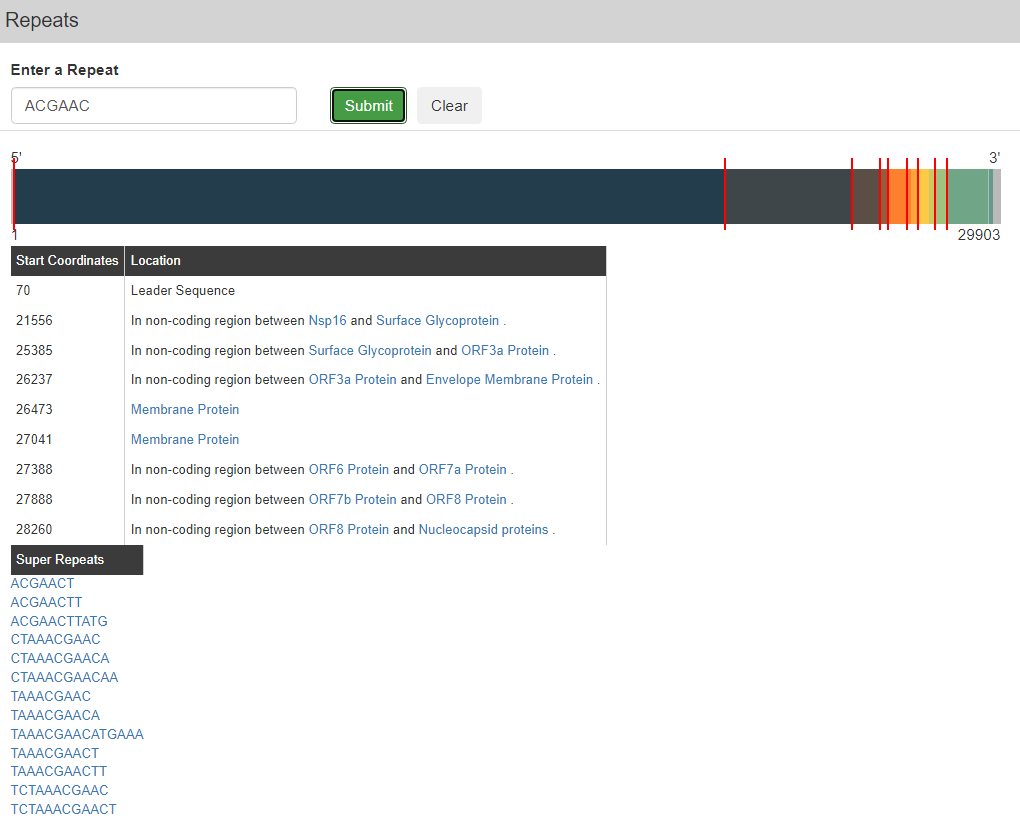
The last option within the “Search” tab is the Repeats option. Once this is right pressed, a page appears where one can input a specific sequence of nucleotides to see specific instances in the viral genome where the sequence occurs. An example is shown below for the “ACGAAC” nucleotide sequence. Once “Submit” is pressed, the system provides a full genome representation with red tick marks of where the repeat occurs. Each occurrence’s start coordinate and location within the genome is provided in a table as shown below and suggestions of existing variations of the inputted repeat are present under “Super Repeats”.